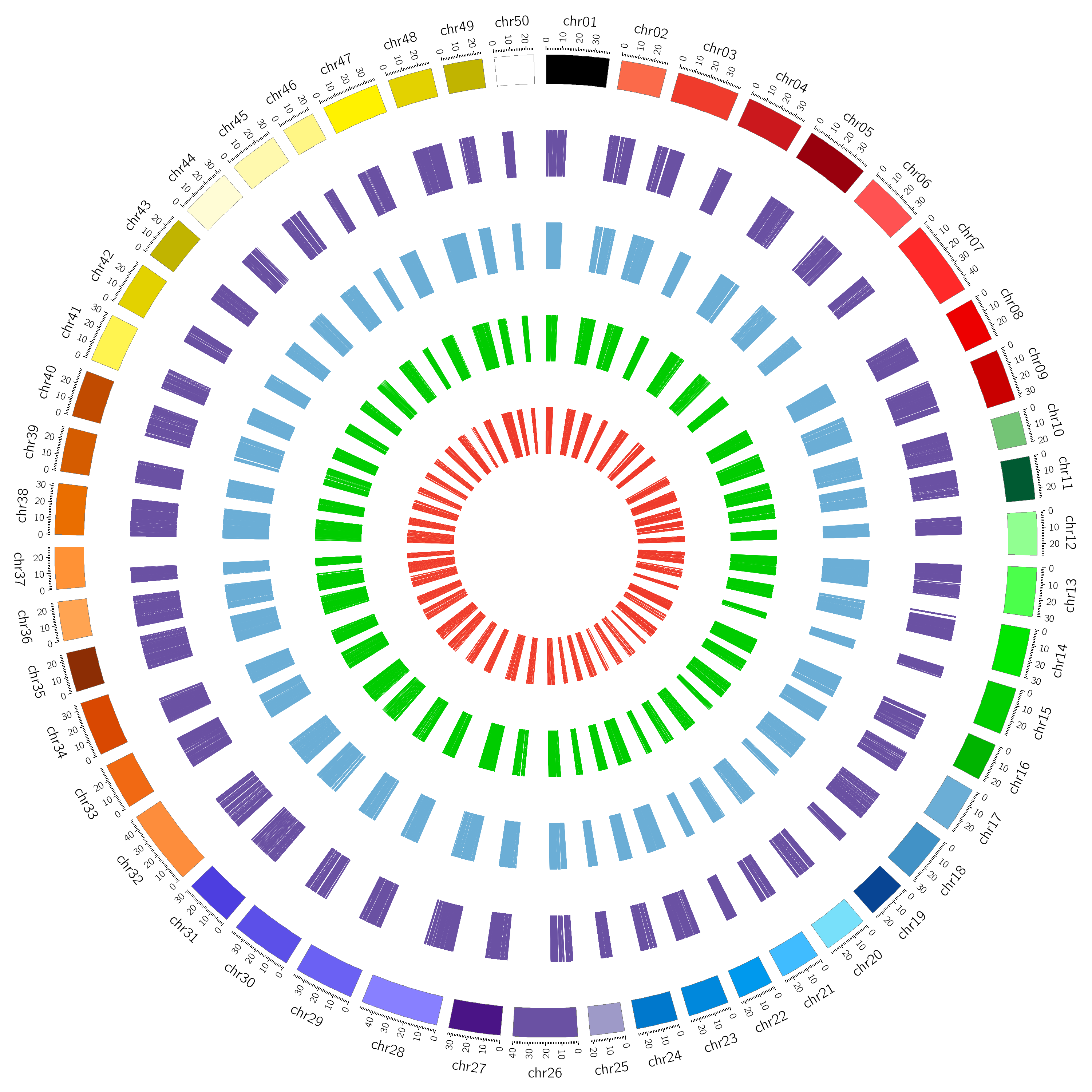

CCncRNAdb (Common Carp non-coding RNA database) is a comprehensive tissue-specific database of Common carp long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) identified from 468 publicly available raw RNA-Seq datasets. Currently, this database has a total of 33,990 lncRNAs and 22,854 circRNAs. The database provides information on the types, chromosomal locations, tissues, lengths of lncRNAs, and circRNAs. Additionally, it provides information on lncRNAs/circRNAs interaction with miRNA and mRNAs. LncRNAs and circRNAs function as competitive endogenous RNAs (ceRNAs), competing with other RNA molecules for shared microRNAs, which ultimately influences gene expression regulation. CCncRNAdb serves as a repository for non-coding RNAs (ncRNAs) in Common carp that are unique to various tissues, such as barb, blood, bone, brain, bulbus, embryo, eye, fin, gallbladder, gill, gonads, gut, heart, hepatopancreas, hypothalamus, intestine, kidney, larvae, liver, muscle, ovary, pituitary, scale, skin, spleen, tail, testis, and thymus. Users could easily access, explore and retrieve all the information related to lncRNAs and circRNAs from this user-friendly database, including downloading, searching, and browsing capabilities. It is our belief that CCncRNAdb is a highly comprehensive and tissue-specific database that will offer new perspectives into the study of lncRNA and circRNA in common carp, ultimately benefiting the wider research community.